
For the Affirmative, David E. Thomas
INTRODUCTION
Evolution is descent with modification - for example, the hypothesized descent of birds from certain theropod dinosaurs. When sons and daughters are born slightly different from their parents (micro-evolution), heredity and descent are involved. "Macro-evolution" means that over time, descent produces new species, genera, families, orders, classes, phyla, and kingdoms.
Charles Darwin had poor models of descent at best. His theory of heredity involved small particles, "gemmules," which flowed through the blood to collect in sexual organs for transmission to the next generation (Darwin 1868). Although Darwin's genetic models were incorrect, he was right about one thing -existing species can evolve into new species.
If this essay were to cover all of the many examples in which molecular comparisons support evolution, it would be thousands of pages long. This evidence could have turned out to be incompatible with common descent in any number of ways. The biomolecular revolution could easily have demolished evolution, exposing it as a "fantasy" based on superficial similarities of a few species. But, again and again, the effects of descent with modification have been confirmed.
During the last century's explosion of biotechnology, molecular biologists have observed only one mechanism for similarity in various organisms' genetic material (DNA) or its expressions (proteins). That mechanism is descent - heredity. A man may protest that he's not the father of a given child, but if that child's DNA has certain similarities to the man's, there is no doubt he is the father. DNA can establish paternity of individuals, and it can also indicate paternity of species - macro-evolution.
Cytochrome c is a vital protein used at the most basic level of cellular electron transport. It is one of a number of ubiquitous proteins - proteins that serve the exact same function in all species. All surviving organisms require a copy of cytochrome c, or an equivalent protein. While these proteins vary from species to species, they all have the same functional shape. Indeed, yeast cells function just fine with human cytochrome c engineered into their cells (Tanaka 1988). There are untold trillions of possible combinations of the 100 or so amino acids involved that make perfectly functional copies of cytochrome c. Only one mechanism has been observed to produce genetic similarity: heredity. Without heredity involved, there is no reason to expect the proteins of any two organisms to be similar.
Evolution demands similarities in the genes and proteins of species thought to have recently evolved from a common ancestor. Since biologists think that humans and chimps both descended from an ancestor who lived just five to eight million years ago, the molecules of inheritance (DNA) and the DNA's expressed proteins should be quite similar between these two species. Conversely, one would expect many more differences between creatures related more distantly, such as humans and turtles.
In fact, there is no difference between the cytochrome c's of human and chimp. Human cytochrome c differs from a rhesus monkey's by just one amino acid, and from an erythrocebus patas monkey's by a different one (Dayhoff 1979). But, humans differ from whales at ten different cytochrome c sites, at 15 for turtles, and so on (Figure 1). There is a "Message" in these proteins: species thought to be closely related turn out to have proteins that are also closely related. If human cytochrome sequences were completely different from those of the apes, or even all other creatures, evolution would have collapsed overnight. Instead, the molecules were in perfect accord with evolutionary expectations - independent and compelling confirmation.
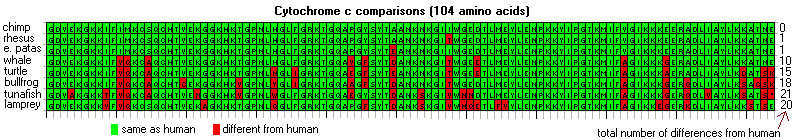
Figure 1 - Comparisons of cytochrome c sequences.
There's an even deeper level of confirmation. It turns out that there are from one to six codons (DNA nucleotide triplets) corresponding to each of the twenty amino acids. There are thus many DNA combinations that can code for the same amino acid sequence. There is no reason to expect that organisms with similar protein sequences should have similar DNA sequences, except for heredity. Figure 2 shows a comparison of human and mice genes and amino acids for cytochrome c. In the absence of a mechanism like heredity, the probability that humans and mice share identical codons for 78 of the 105 amino acids in the protein is astonishingly small: 7.6*10-34 (NCBI 2002, Hinrichs 2000, Theobald 2002).
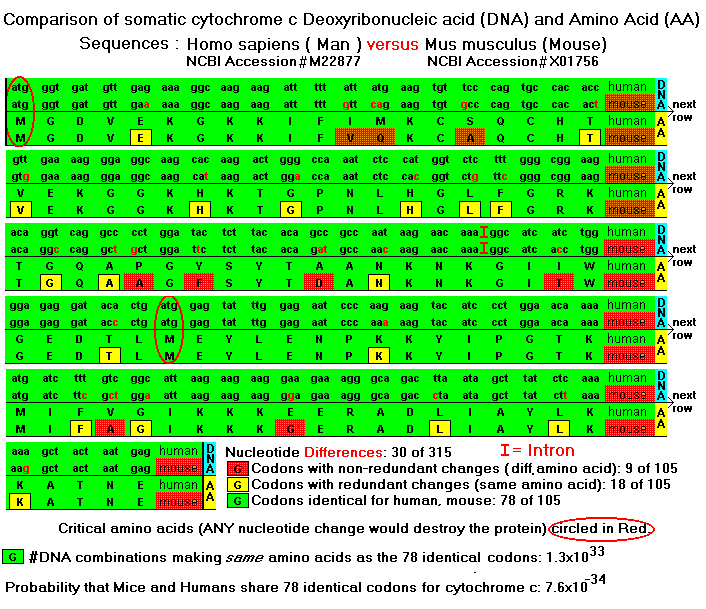
Figure 2. DNA Redundancies in Man and Mouse
Cytochrome c phylogenies are sometimes imperfect, since they are based on a single gene, and mutations can occasionally reverse previous mutations. In Figure 1, the lamprey appears closer to humans than to tuna fish, contrary to evolutionary expectations. Biologists have addressed this problem by looking at more and more genes. Some genes evolve faster than others, some slower. Of course, the fact that some genes change faster or slower than others doesn't disprove evolution.
A recent study (Kumar and Hedges 1998) involved hundreds of genes, for dozens of species. In Figure 3, the 50 genes used to compare old world monkeys to humans have a strong "peak" indicating 23 million years (MYa) since the last common ancestor of these species. The bell-shaped curve centered at 23 MYa is nothing less than a strong biotic "signal." When 107 genes of primates and rabbits are compared, these differences correspond to 90 MYa since the primate/rabbit common ancestor. The "message" is clear: humans are genetically much closer to old world monkeys than they are to rabbits. This accords perfectly with evolutionary expectations: the last common ancestor of primates and rabbits is much farther back in time than the last common ancestor of humans and old world monkeys. Had the genes turned out otherwise, Darwin's "fantasy" would have evaporated.
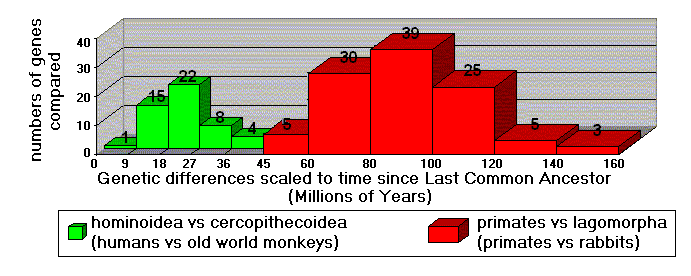
Figure 3. Comparisons of hundreds of genes. (Data from Kumar & Hedges1998)
Most organisms have the very same genetic code, the association of DNA/RNA triplets with specific amino acids. For example, in most organisms, UGA is a "stop" codon, and specifies the end of a protein. But in some Mycoplasma bacteria, UGA codes for the amino acid tryptophan instead (Inamine 1990). Several natural hypotheses for the evolution of non-standard genetic codes have been advanced and published (Osawa and Jukes 1995). If all creatures are descended from one or a few ancestors, then such variant genetic codes should also display a pattern of derived similarities and differences, just like those in protein sequence comparisons. And so they do (Knight 2001, Miller 2001). The real significance of the few non-standard code exceptions is that they demonstrate that organisms can indeed exist with their own unique genetic code translations. There is nothing to stop a Designer from giving any species such as humans a special genetic code, unlike that of any other creature (Hofstadter 1982). The 2.3*1069 possible genetic codes are more than enough for each baramin (kind), even each species, to have its own special code. This, again, could have dramatically falsified evolution.
Some of the most compelling molecular evidence for evolution involves inheritance of non-functional DNA, like introns (Figure 2). While most vertebrates synthesize their own vitamin C (ascorbic acid), some primates can't. When sequences of vertebrates are compared, we find that primates do possess genes for making vitamin C. In humans and apes, however, these genes are non-functional. A mutation has destroyed the "recipe," but it's not a "harmful" mutation because primates get vitamin C in their diets. The mutations blocking human production of this enzyme are identical to those in the apes. It's unlikely the same mistakes happened independently, in exactly the same positions; it's more likely that humans and apes inherited this error from their common ancestor. (Max 2000). Additionally, curious DNA fragments called "transposons" ("jumping genes") can inject their DNA randomly into the genome (Armstrong 2000). Once inserted, these genetic fragments are then inherited. Some SINE (short interspersed elements) transposons are routinely used for paternity suits (Novick 1993, 1995); others show the same genetic elements, randomly inserted into the same locations, in both humans and chimps (Sawada 1985).
Nylon wasn't invented until 1937. When scientists found a new species of bacteria feasting on nylon byproducts, they became quite interested. When they compared old and new strains, they found the new strain must have nylon byproducts to live, and that its novel nylon-digesting abilities were due to a frame shift mutation in one gene. In a frame shift, a mutation of just one nucleotide can push the whole sequence by a notch, creating a brand-new protein. In this case, the protein reacted to nylon products, and suddenly gave the bacteria a whole new niche. The novel proteins we see in real life do indeed appear to be due to inherited changes in DNA, just as evolution demands (Ohno 1984, Thomas 2002).
In each of these cases, things could have turned out in thousands of different ways, each with the potential to disprove evolution. But that's not what happened. Comparisons of molecules provide independent and compelling support for macro-evolution.
W.P. Armstrong, (2000). "Transposons (Jumping Genes)," http://waynesword.palomar.edu/transpos.htm.
C. R. Darwin (1868). The Variation of Animals and Plants under Domestication , ISBN: 0801858674.
Margaret Dayhoff (1979) Atlas of Protein Sequence and Structure, ISBN: 0912466073; (April 1979 edition)
Steve Hinrichs (June 2000). "Compelling Data for Common Descent from Matching Redundant DNA Sequences," http://members.aol.com/SHinrichs9/descent/descent.htm.
Douglas Hofstadter (1982). "Is the genetic code an arbitrary one, or would another code work just as well?", Scientific American, March 1982, 18-29.
J.M. Inamine et. al. (1990). "Evidence that UGA is read as tryptophan rather than stop by Mycoplasma ...," Journal of Bacteriology 172;504-506.
Robin Knight et. al., (2001) "Rewiring The Keyboard: Evolvability Of The Genetic Code," Nature Reviews - Genetics. 2: 49-58.
S. Kumar and S. Hedges (1998). "A molecular timescale for vertebrate evolution," Nature, Vol. 392, 30 April 1998, pp. 917-920.
Edward Max (2000). "Plagiarized Errors and Molecular Genetics," <http://www.talkorigins.org/faqs/molgen/>.
Kenneth R. Miller (2001), http://www.ncseweb.org/resources/articles/6945_km-3.pdf.
NCBI (2002). "Somatic cytochrome c Gene," Accession Number M22877 (Human), X01756 (mouse).
G. E. Novick, et. al., (1993). "The use of polymorphic Alu insertions in human DNA fingerprinting," EXS. 67: 283-291. [PubMed]
G. E. Novick, et. al., (1995) "Polymorphic human specific Alu insertions ...," Electrophoresis 16: 1596-1601. [PubMed]
Susumu Ohno (1984) "Birth of a unique enzyme ...," Proc. Natl. Acad. Sci. USA, Vol. 81, pp. 2421-2425, April 1984.
S. Osawa, T. H. Jukes (1995) "On Codon Reassignment," Journal of Molecular Evolution, 41:247-249.
I. Sawada, et al. (1985) "Evolution of Alu family repeats since the divergence of human and chimpanzee," Journal of Molecular Evolution 22(316). [PubMed]
Y. Tanaka, et. al. (1988) "Construction of a human cytochrome c gene and its functional expression in Saccharomyces cerevisiae," J Biochem (Tokyo) (1988) 103: 954-61.[PubMed]
Douglas Theobald (2002). "The Molecular Sequence Evidence," http://www.talkorigins.org/faqs/comdesc/section4.html.
David E. Thomas (July 2002) "Evolution and Information: The Nylon Bug," http://www.nmsr.org/nylon.htm.
To contact Mr. Thomas, click HERE.